Part 2: On practicing what we preach Originally posted on Medium. A few weeks ago I described the results of a project investigating the data management…
Tag: open data
By Daniella Lowenberg “Listening is not merely not talking, though even that is beyond most of our powers; it means taking a vigorous, human interest…
Neuroimaging as a case study in research data management: Part 1
Posted in Digital Curation, and Projects
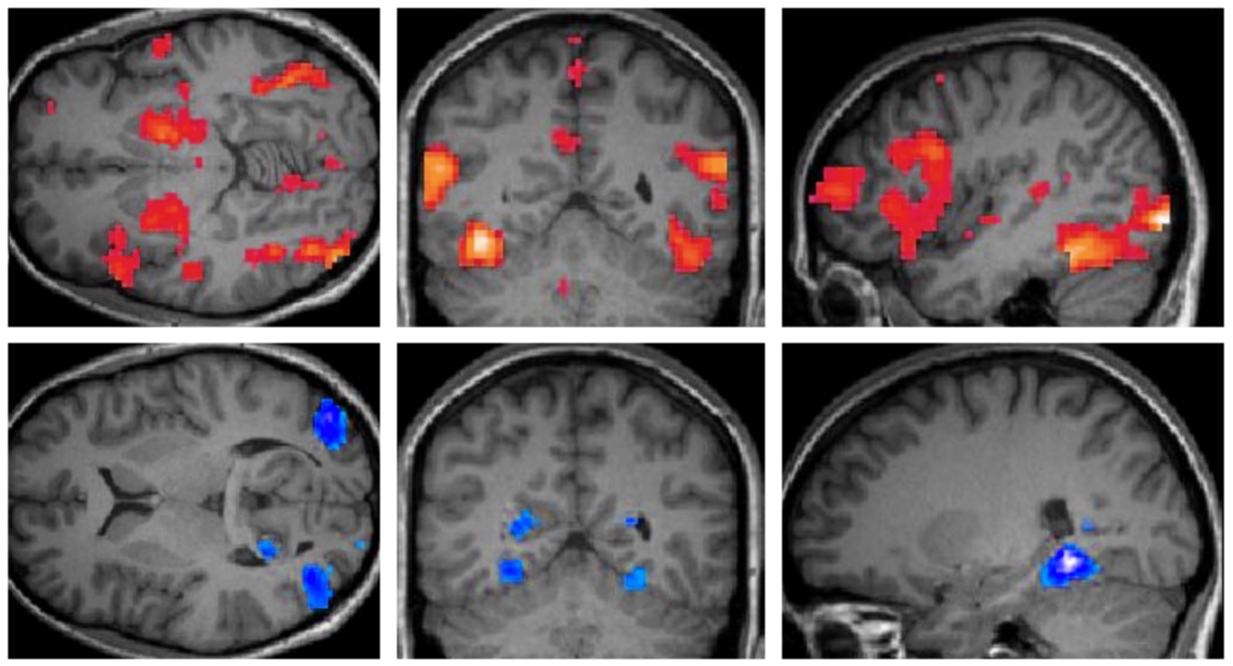
Part 1: What we did and what we found This post was originally posted on Medium. How do brain imaging researchers manage and share their…
Support your Data
Posted in Uncategorized
Building an RDM Maturity Model: Part 4 By John Borghi Researchers are faced with rapidly evolving expectations about how they should manage and share their data,…
Dash: 2017 in Review
Posted in Data Publication, and Development
The goal for Dash in 2017 was to build out features that would make Dash a desirable place to publish data. While we continue to…
Test-driving the Dash read-only API
Posted in Data Publication, and Development
The Dash Data Publication service is now allowing access to dataset metadata and public files through a read-only API. This API focuses on allowing metadata…
By Daniella Lowenberg At RDA10 in Montreal I gave a presentation on Dash in the Repository Platforms for Research Data IG session. The session was…
Dash Updates: Fall, 2017
Posted in Data Publication
Throughout the summer the Dash team has focused on features that better integrate with researcher workflows. The goal: make data publishing as easy as possible.…
By John Borghi and Daniella Lowenberg Happy Friday! This week we’ve defined open data, discussed some notable anecdotes, outlined publisher and funder requirements, and described…
By John Borghi and Daniella Lowenberg Yesterday we talked about about why researchers may have to make their data open, today let’s start talking about…